OBJECTIVES
– To test whether stimulated B cells differentially express IL-10 and/or Fcgr2b genes upon IVIg treatment.
MATERIALS
– High Capacity RNA-to-cDNA Kit (Ref:4387406, Applied Biosystems)
– TaqMan Universal Master Mix II with UNG (Ref:4440042, Applied Biosystems)
– Human IL-10 Assay Hs00961622-m1 (Applied Biosystems)
– Human Fcgr2b Assay Hs01634996-s1 (Applied Biosystems)
– Human GAPDH Assay Hs99999905-m1 (Applied Biosystems)
– Human 18S Assay Hs99999901-s1 (Applied Biosystems)
PROTOCOLS
A) RT (20161104)
Samples were retrotranscribed according to the following protocol. A blank sample (w/o RNA template) as a negative control was added to the reaction.
| n=1 | MM (n=18+2=20) | |
|---|---|---|
| 2X RT Buffer | 10 mcL | 200 mcL |
| 20X RT Enzyme mix | 1 mcL | 20 mcL |
| Nuclease-free H2O | qsp. 20 mcL | - |
| Sample | up to 9 mcL | - |
| Total per reaction | 20 mcL | Add 11 mcL per microtube |
200 ng of sample RNA were used in each 20 mcLreaction. The following volumes were added per microtube.
| Datos | TTO | Cuantificación RNA | Volumen muestra RT (200 ng) | Volumen H2O RT (qsp 9 mcl) | ||
|---|---|---|---|---|---|---|
| Nº individuo | Nombre | IVIg | Concentración (ng/mcL) | mcL | mcL | |
| 1 | Cinta | NO | 88,7 | 2.2547914317926 | 6.7452085682074 | |
| SI | 114,3 | 1.7497812773403 | 7.2502187226597 | |||
| 2 | Elena | NO | 186,7 | 1.0712372790573 | 7.9287627209427 | |
| SI | 89,4 | 2.2371364653244 | 6.7628635346756 | |||
| 3 | Jordi | NO | 223,8 | 0.89365504915103 | 8.106344950849 | |
| SI | 198,7 | 1.0065425264217 | 7.9934574735783 | |||
| 4 | Noemi | NO | 150,6 | 1.32802124834 | 7.67197875166 | |
| SI | 114,6 | 1.7452006980803 | 7.2547993019197 | |||
| 5 | Tomas | NO | 109,3 | 1.8298261665142 | 7.1701738334858 | |
| SI | 105,4 | 1.8975332068311 | 7.1024667931689 | |||
| 6 | Ricard | NO | 134,8 | 1.4836795252226 | 7.5163204747774 | |
| SI | 130,5 | 1.5325670498084 | 7.4674329501916 | |||
| 7 | Xavi | NO | 159,9 | 1.2507817385866 | 7.7492182614134 | |
| SI | 246,8 | 0.81037277147488 | 8.1896272285251 | |||
| 9 | Celia | NO | 151.4 | 1.3210039630119 | 7.6789960369881 | |
| SI | 176.2 | 1.1350737797957 | 7.8649262202043 | |||
| 10 | Nacho | NO | 211.2 | 0.9469696969697 | 8.0530303030303 | |
| SI | 164.4 | 1.2165450121655 | 7.7834549878346 |
The thermal cycler conditions were as follows, as indicated by Applied (total volume 20 mcL):
| Step 1 | Step 2 | Step 3 | |
|---|---|---|---|
| Temperature (º C) | 37 | 95 | 4 |
| Time | 60 min | 5 min | Infinite Hold |
C) qPCR (20161107)
Samples were analized in triplets and two blank samples were added to the reaction as negative controls: one sample w/o cDNA template (qPCR blank) and the RT negative control.
The following protocol was used:
| N=1 | MM per detector (n=54+3) | |
|---|---|---|
| Taqman Universal Master Mix II | 10 mcL | 570 mcL |
| Taqman Gene Expression Assay | 1 mcL | 57 mcL |
| cDNA template+ H2O | up to 9 mcL* | - |
| Total volume | 20 mcL | Add 11 mcL per well |
0,6 mcL of sample (200ng/20 mcL*0,6 mcL=6 ng) and 8,4 mcL of water were added per well. Since 12 wells (n=12+2=14) were analized per sample, a mastermix of 8,4 mcl of sample+ 117,6 mcl of water.
The thermal cycler conditions were as follows, as indicated by Applied (total volume 20 mcL):
| UNG incubation (Hold) | Polymerase activation (Hold) | PCR (40 cycles) | |
|---|---|---|---|
| Temperature (ºC) | 50 | 95 | 95 + 60 |
| Time (mm:ss) | 2:00 | 10:00 | 00:15 + 1:00 |
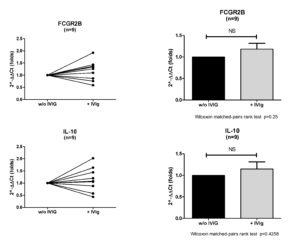
RESULTS
CONCLUSIONS
Stimulated B cells do not differentially express IL-10 or Fcgr2b genes upon IVIg treatment.